Introduction
Introduction¶
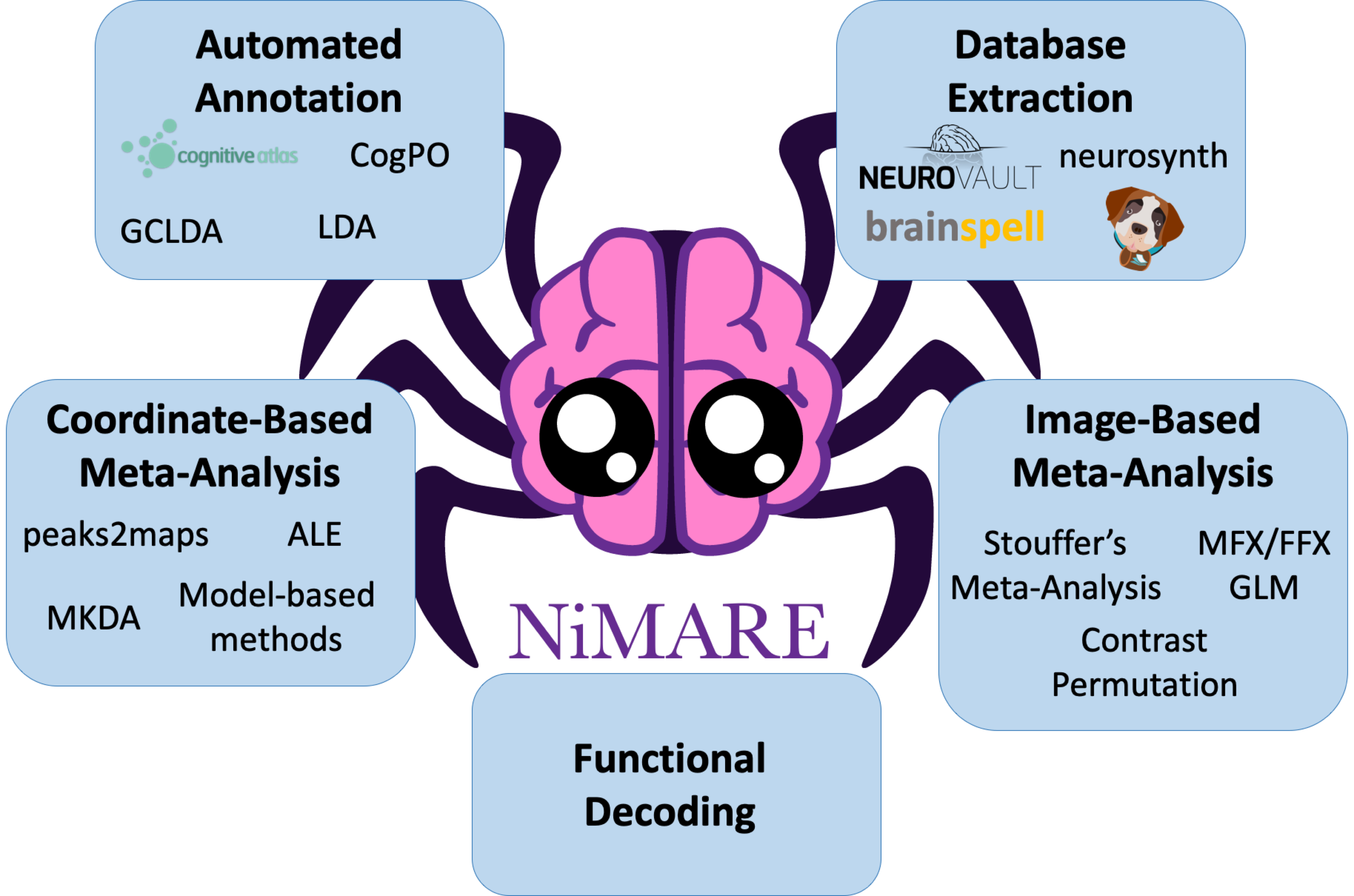
We introduce NiMARE (Neuroimaging Meta-Analysis Research Environment), a Python package for analyzing meta-analytic neuroimaging data. NiMARE is a new library developed as a component in a burgeoning open-source meta-analytic ecosystem for neuroimaging data, which currently includes Neurosynth, NeuroVault, NeuroQuery, and PyMARE.
While several libraries already exist for neuroimaging meta-analysis, these libraries are generally algorithm-specific, and are provided in a range of very different user interfaces, languages, and licenses. This variability may prevent meta-analysts from using the most appropriate algorithm for a given analysis. Further, having multiple meta-analysis algorithms available in one library facilitates direct comparisons of methods. With NiMARE, we consolidate meta-analytic algorithms from a range of libraries and publications, and provide a common Python syntax and well documented application program interfaces. Additionally, NiMARE is a collaboratively-developed open source package, enabling researchers to contribute new methods not included in the current version.
In this paper, we describe NiMARE’s aims, architecture and the functionality it supports—including tools for database extraction, automated annotation, meta-analysis, meta-analytic coactivation modeling, and functional decoding. The text is accompanied by extensive code samples and results (also available online in the form of Python scripts; https://github.com/NBCLab/nimare-paper with additional documentation in https://github.com/neurodatascience/meta_analysis_notebook), ensuring that users can follow along interactively.