NiMARE Overview
Contents
NiMARE Overview¶
NiMARE is designed to be modular and object-oriented, with an interface that mimics popular Python libraries, including scikit-learn and nilearn. This standardized interface allows users to employ a wide range of meta-analytic algorithms without having to familiarize themselves with the idiosyncrasies of algorithm-specific tools. This lets users use whatever method is most appropriate for a given research question with minimal mental overhead from switching methods. Additionally, NiMARE emphasizes citability, with references in the documentation and citable boilerplate text that can be copied directly into manuscripts, in order to ensure that the original algorithm developers are appropriately recognized.
NiMARE works with Python versions 3.6 and higher, and can easily be installed with pip.
Its source code is housed and version controlled in a GitHub repository at https://github.com/neurostuff/NiMARE.
NiMARE is under continued active development, and we anticipate that the user-facing API (application programming interface) may change over time. Our emphasis in this paper is thus primarily on reviewing the functionality implemented in the package and illustrating the general interface, and not on providing a detailed and static user guide that will be found within the package documentation.
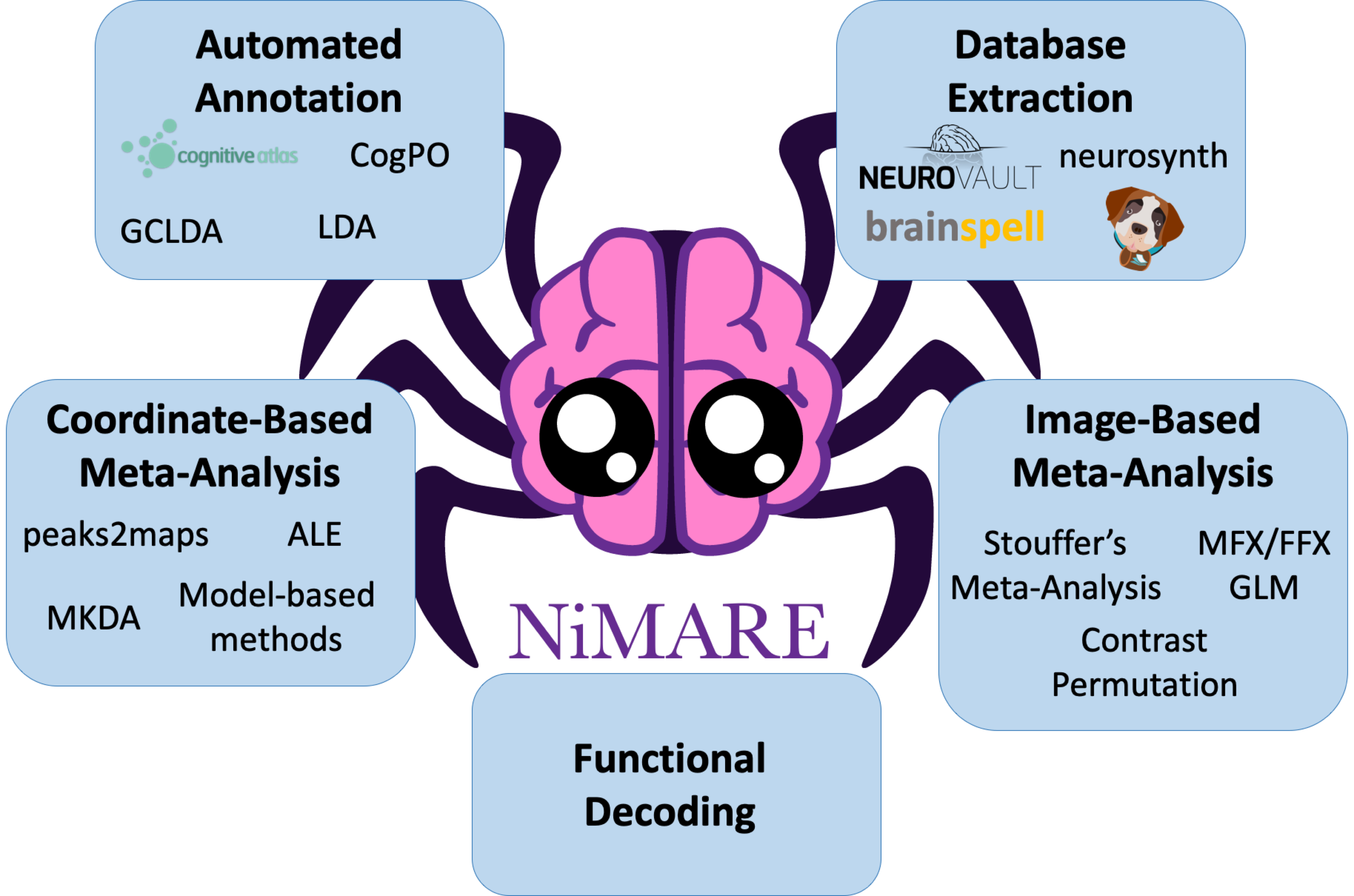
Tools in NiMARE are organized into several modules, including nimare.meta, nimare.correct, nimare.annotate, nimare.decode, and nimare.workflows.
In addition to these primary modules, there are several secondary modules for data wrangling and internal helper functions, including nimare.io, nimare.dataset, nimare.extract, nimare.stats, nimare.utils, and nimare.base.
These modules are summarized in Application Programming Interface, as well as in Table 1.
Application Programming Interface¶
One of the principal goals of NiMARE is to implement a range of methods with a set of shared interfaces, to enable users to employ the most appropriate algorithm for a given question without introducing a steep learning curve.
This approach is modeled on the widely-used scikit-learn package [Pedregosa et al., 2011, Buitinck et al., 2013], which implements a large number of machine learning algorithms - all with simple, consistent interfaces.
Regardless of the algorithm employed, data should be in the same format and the same class methods should be called to fit and/or generate predictions from the model.
To this end, we have adopted an object-oriented approach to NiMARE’s core API that organizes tools based on the type of inputs and outputs they operate over.
The key data structure is the Dataset class, which stores a range of neuroimaging data amenable to various forms of meta-analysis.
There are two main types of tools that operate on a Dataset class.
Transformer classes, as their name suggests, perform some transformation on a Dataset- i.e., they take a Dataset instance as input, and return a modified version of that Dataset instance as output (for example, with newly generated maps stored within the object).
Estimator classes apply a meta-analytic algorithm to a Dataset and return a set of statistical images stored in a MetaResult container class.
The key methods supported by each of these base classes, as well as the main arguments to those methods, are consistent throughout the hierarchy (e.g., all Transformer classes must implement a transform() method), minimizing the learning curve and ensuring a high degree of predictability for users.
Fig. 2 A schematic figure of Datasets, Estimators, Transformers, and MetaResults in NiMARE.¶
Package Organization¶
At present, the package is organized into 14 distinct modules.
nimare.dataset defines the Dataset class.
nimare.meta includes Estimators for coordinate- and image-based meta-analysis methods.
nimare.results defines the MetaResult class, which stores statistical maps produced by meta-analyses.
nimare.correct implements Corrector classes for family-wise error (FWE) and false discovery rate (FDR) multiple comparisons correction.
nimare.annotate implements a range of automated annotation methods, including latent Dirichlet allocation (LDA) and generalized correspondence latent Dirichlet allocation (GCLDA).
nimare.decode implements a number of meta-analytic functional decoding and encoding algorithms.
nimare.io provides functions for converting alternative meta-analytic dataset structure, such as Sleuth text files or Neurosynth datasets, to NiMARE format.
nimare.transforms implements a range of spatial and data type transformations, including a function to generate new images in the Dataset from existing image types.
nimare.extract provides methods for fetching datasets and models across the internet.
nimare.generate includes functions for generating data for internal testing and validation.
nimare.base defines a number of base classes used throughout the rest of the package.
Finally, nimare.stats and nimare.utils are modules for statistical and generic utility functions, respectively.
These modules are summarized in Table 1.
Module |
Description |
|---|---|
|
This module stores the |
|
This module contains |
|
This module stores the |
|
This module contains classes for multiple comparisons correction, including |
|
This module includes a range of tools for automated annotation of studies. Methods in this module include: topic models, such as latent Dirichlet allocation and generalized correspondence latent Dirichlet allocation; ontology-based annotation, such as Cognitive Atlas term extract from text; and general text-based feature extraction, such as count or tf-idf extraction from text. |
|
This module includes a number of methods for functional characterization analysis, also known as functional decoding. Methods in this module are divided into three groups: discrete, for decoding regions of interest or subsets of the Dataset; continuous, for decoding unthresholded statistical maps; and encoding, for simulating statistical maps from labels. |
|
This module contains functions for converting common file types, such as Neurosynth- or Sleuth-format files, into NiMARE-compatible formats, such as |
|
This module contains classes and functions for converting between common data types. Two important classes in this module are the |
|
This module contains functions for downloading external resources, such as the Neurosynth dataset and the Cognitive Atlas ontology. |
|
This module contains miscellaneous statistical methods used throughout the rest of the library. |
|
This module contains functions for generating useful data for internal testing and validation. |
|
This module contains miscellaneous utility functions used throughout the rest of the library. |
|
This module contains a number of common workflows that can be run from the command line, such as an ALE meta-analysis or a contrast-permutation image-based meta-analysis. All of the workflow functions additionally generate boilerplate text that can be included in manuscript methods sections. |
|
This module defines a number of base classes used throughout the rest of the library. |
Dependencies¶
NiMARE depends on the standard SciPy stack, as well as a small number of widely-used packages.
Dependencies from the SciPy stack include scipy [Virtanen et al., 2020], numpy [Walt et al., 2011, Harris et al., 2020], pandas [McKinney, 2010], and scikit-learn [Pedregosa et al., 2011, Buitinck et al., 2013].
Additional requirements include fuzzywuzzy, nibabel [Brett et al., 2020], nilearn [Abraham et al., 2014], statsmodels [Seabold and Perktold, 2010], and tqdm [da Costa-Luis et al., 2020].